Snippets 17
The ubiquitous universally Conserved Sequence Block 3 (CSB-3)
Alice C. Lichtenstein, M.S.
aclsnippets@gmail.com
Introduction
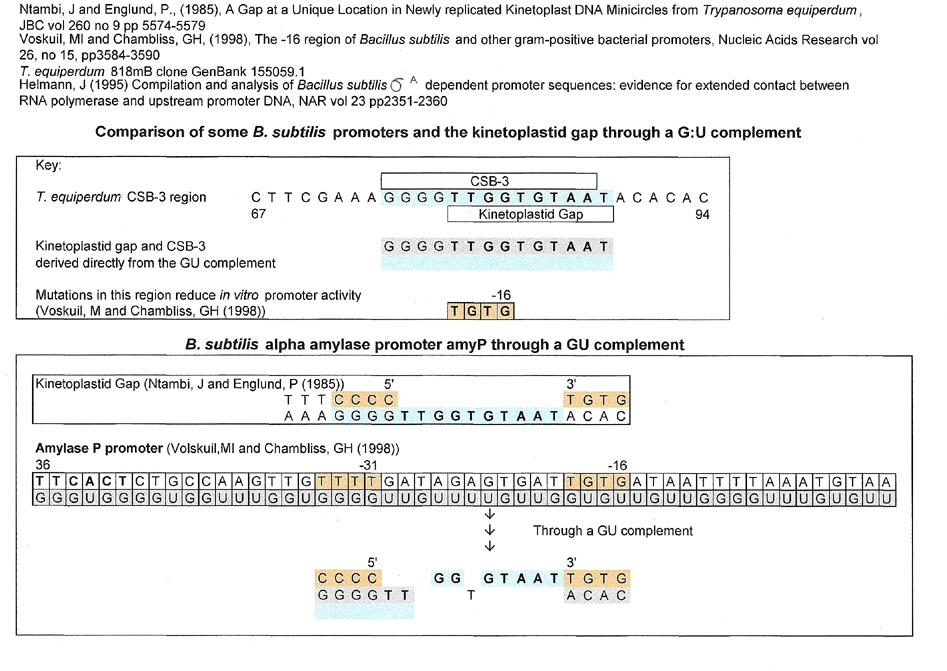
All kinetoplast mitochondrial minicircles have a universally conserved sequence block (Conserved Sequence Block 3 or CSB-3) (GGGGTTGGTGTA). In T. equiperdum, there is a smaller sequence, the kinetoplastid gap (TTGGTGTAAT) that overlaps and is partially embedded in CSB-3. Judging from the online literature, the kinetoplastid gap was first described by Ntambi, J and Englund, P (1985) as a single stranded region in a newly replicated kinetoplastid double-stranded minicircle. In their next paper, Ntambi, J and Englund, P (1986) reported that they found ribonucleotides covalently linked to the 5' end of the new strand in the gap, "perhaps the remnants of a replication primer".
from fig. 6 Ntambi, J and Englund P (1985):
| 5' | Kinetoplastid Gap | 3' | |||||||||||||||||
| T | T | T | C | C | C | C | T | G | T | ||||||||||
| A | A | A | G | G | G | G | T | T | G | G | T | G | T | A | A | T | A | C | A |
|
Conserved Sequence Block 3 (CSB-3) |
|||||||||||||||||||
CSB-3 is found, without variation in thousands of kinetoplastid mitochondrial minicircles. The original kinetoplastid gap (Ntambi, J and Englund, P (1986)) (TTGGTGTAAT) was found using Trypanosma equiperdum and while the gap sequence can be found in other trypanosomes (e.g T. brucei), none have been described in as much detail as the one from T. equiperdum.. A body of work by Shlomai, J's laboratory (Abu-Einell et al. (1999)) has shown, using Crithidia fasciculata that the CSB-3 sequence (GGGGTTGGTGTA) and a hexameric sequence (ACGCCC) are probably binding sites for what they call an "initiator protein", the universal minicircle sequence binding protein (UMSBP). This protein is associated with minicircle replication; lending support to the notion that the kinetoplastid gap serves as an RNA primer site for replication. For our purposes, this installment of Snippets will only be looking for CSB-3 and the kinetoplastid gap, not the hexameric sequence.
While comparing many sequences using a G:U intermediate, it was found that many matches (see below under methods how a match is made) turned out to be the kinetplastid gap sequence, if not the complete CSB-3 sequence. Some of these matches have been reported in previous Snippets installments on this website and others will be new. This installment will therefore have the quality of a list, but I thought it important enough to show the diversity of locations where CSB-3 and kinetoplastid gap matches were found.
Unfortunately, and confusingly, for a given sequence of interest, there were many more than one CSB-3 or kinetoplast match...some going 5'-3', others going 3'-5',some derived from the G:U complement itself, and some overlapping; all these results with no discernable trend or correlations to anything. This author's reaction was "Oh dear, no one is going to believe any of these results in Snippets as other than my fertile 'best fit' imagination".
Spoiler alert: after staring at all the matches for a while it became apparent that there was an underlying triplet sequence that gave rise to the many results. It is unclear what the triplet sequence was, but looking through a G:U intermediate, this author chose (CAG)n. (TAA)n might have been a good choice too, but it really didn't matter.
So, let us begin with Methodology and how matches were made.
Methods
To begin with, all the nucleotide sequences used were considered as 'strings of beads" that had no other meaning, for the present, as to function or location, or species etc. They were analyzed both in the 5-'3' and 3'-5' direction.
An uncommon nucleotide like di-hydroxyuridine (D) was considered to be uridine because the only sequence interaction used in this analysis was complementation.
T and U were used interchangeably.
Proteins were not considered in any of the methods.
The method used to compare sequences of interest with known conserved sequences was done similarly to the anti-PAS binding site sequence described below. As an example, to create a G:U intermediate , a complement of G's and Us (gray background) was generated from a known sequence of interest such as the anti-PAS binding site (CCAGGG) located in tRNA-lysine3 (UUU), a retroviral primer, (The anti-PAS binding site is being used as an example of the method, ).
Generating a G:U Complement
| Anti-pas sequence of Retroviral primer | C | C | A | G | G | G |
| G:U complement | G | G | U | U | U | U |
| Reverse complement | C or T or U | C or T or U | A or G | A or G | A or G | A or G |
Using this methodology, some of the following "matches" for CCAGGG (one of the binding sites of tRNAlysine3 primer to a retrovirus template) could be made
| Anti-pas sequence of Retroviral primer | C | C | A | G | G | G |
| G:U complement | G | G | U | U | U | U |
| homology | C | C | A | G | G | G |
| match | T | C | A | G | G | G |
| Telomere rpt humans, mice | T | T | A | G | G | G |
| match | C | C | G | G | G | G |
| match | C | T | A | G | G | G |
| match | C | T | G | G | G | G |
| Telomere rpt. Tetrahymena | T | T | G | G | G | G |
| match to G:U complement | G | A | T | T | T | T |
| match to G:U complement | A | A | C | C | T | C |
| match to both | T | T | T | T | T | T |
The fact that a match could be found between the retroviral primer anti-pas site and the human and tetrahymena telomere repeat did and does not mean that there is any known biological or causal relationship between the two.
It is important to note that a match can be made to both the positive strand (the sequence of interest) and its G:U complement (see last line of above chart of results). This author made the arbitrary decision that the split sections of a match made between the both the sequence of interest and its G:U complement had to be at least 4 or 5 nucleotides in length.
In essence, one is substituting a purine for another purine or a pyrimidine for another pyrimidine to make a match. One could use the letters "Y" for pyrimidine or "R" for purine instead of G and U but this author liked G and U. Even the numbers 1 or 0 could be used provided they were each defined as a specific set of nucleotide bases (ie 1 stands for A or G and 0 stands for C, U, or T).
Unlike homologies, there can be more than one match for any given sequence of interest. This is important.
One author, working with homologies used an 80% similarity as a criteria, and this author has kept that at the back of her mind when subjectively making matches.
This author used a number of papers from the 1980s because she found that using large genomic databases had the following problem. She would locate a sequence in the large database and then when she went back to find it, the database had been updated and the sequence was not at the same location. (The nucleotide numbers were different). So, if the database had a manuscript reference for the original sequence and the original sequence was still the same as the one in the database she used the original paper as a reference
The complete sequence of Trypanosoma equiperdum clone 818mB minicircle, (GenBank EU155059.1) was used for comparisons. Other T. equiperdum clones had the same sequences around the CSB-3 region, but their base numbers were different, so the 818mB minicircle clone was used for all comparisons in this installment of Snippets.
Results and Discussion
Rather than elaborate on the sequences used for comparisons and the content of each article this author has used a quote from the article or maybe one or two sentences as an explanation of the sequences of interest. The references for each comparison are listed above the illustrations in the graphic if the reader wants more information or clarification.
The sequences of interest that were studied for matches to T. equiperdum's CSB-3 region were
Maize mitochondrial conserved core element
Group II intron insertion site
Telomere repeats, human mu switch region
Bacterial promoters were the first set of sequences that were searched for matches with CSB-3 and the kinetoplastid gap.

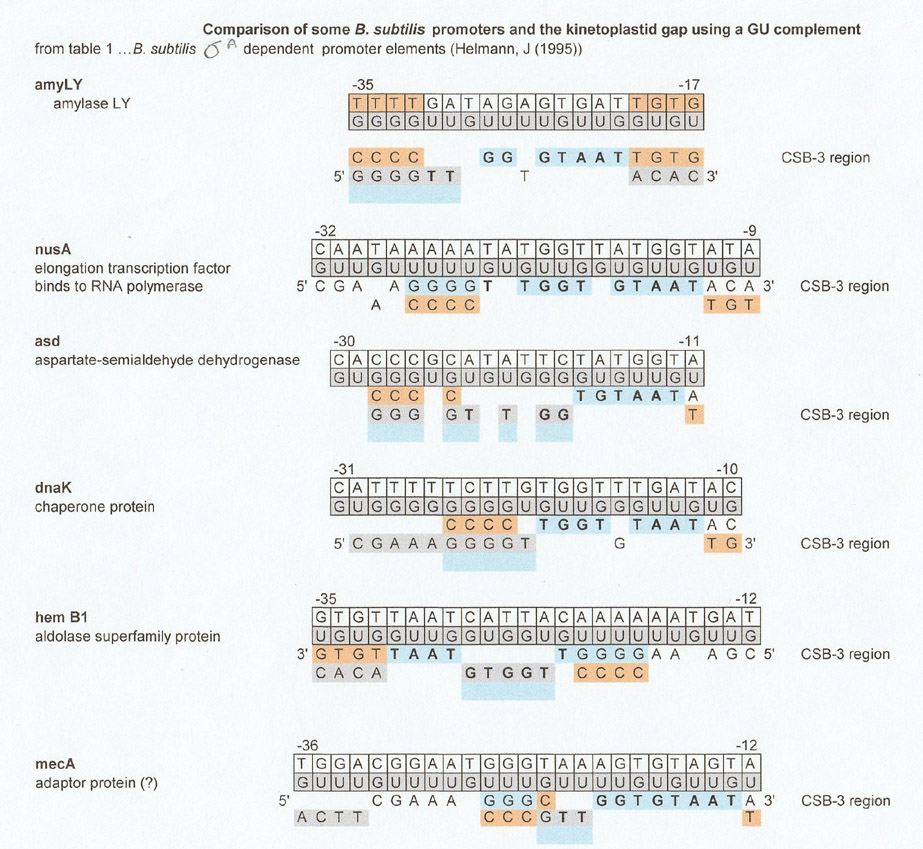
The matches with the kinetoplastid gap show that its orientation can be 5' to 3', 3' to 5' and divided between the sequence of interest (the promoter) and its G:U complement.
The above matches between some B. subtilis promoters and the kinetoplastid gap sequence looked tenable, but disappointingly many other matches to the CSB-3 region could also be found for any given promoter sequence. The question arose then, were the numerous matches simply a failing of the method used, in other words an over aggressive "best-fit" analysis or did they reflect something inherent in the promoter sequence itself? This also brought up another weakness in the matches, there was no control. After pondering this for a few days, this author figured that there might be an underlying shorter repeat sequence that would account for all the matches. The question was, which triplet repeat. For our findings, it didn't matter, so she chose (CAG)n, but through a GU complement, it could have been (UGG)n, (TAA)n or any YRR or RYY triplet repeat except for RRR or YYY. More on this topic later.
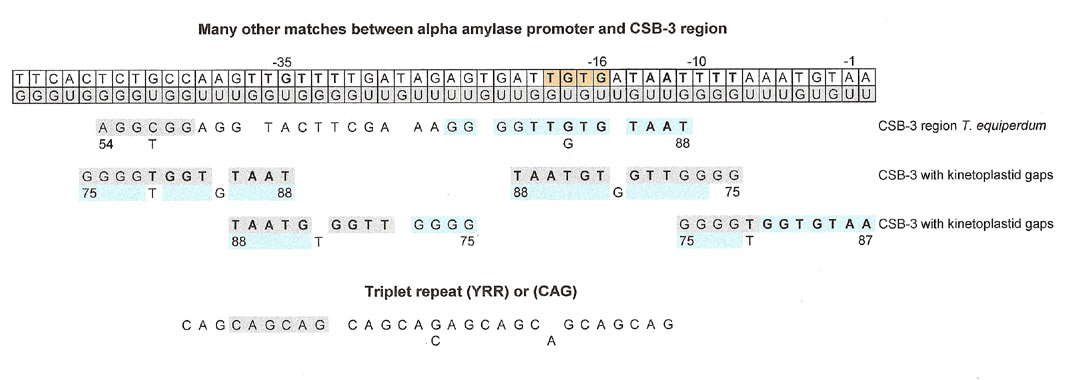
Also, not all the studied promoter sequences had a match for the CSB-3 region or kinetoplastid gap. This author didn't look for other GU- base patterns to see if she could find other conserved sequences (primers?) that would be consistent matches for the remaining promoters.
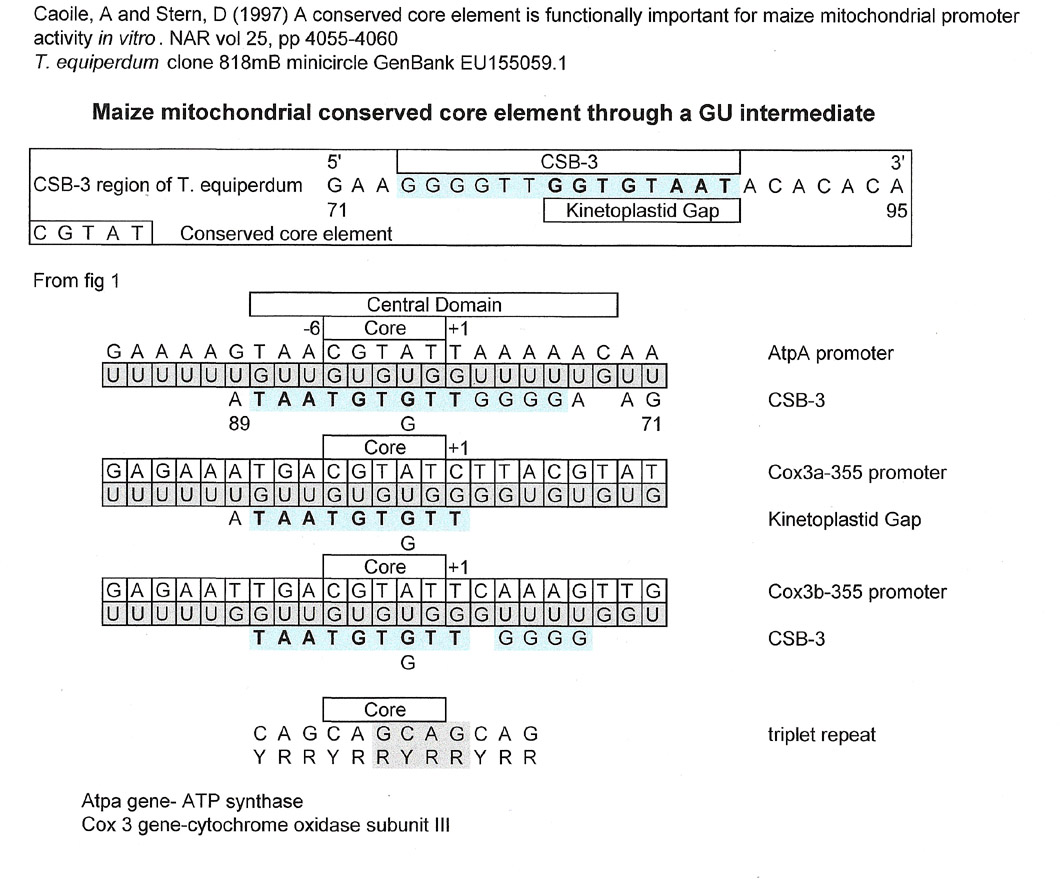
Maize mitochondrial conserved core element
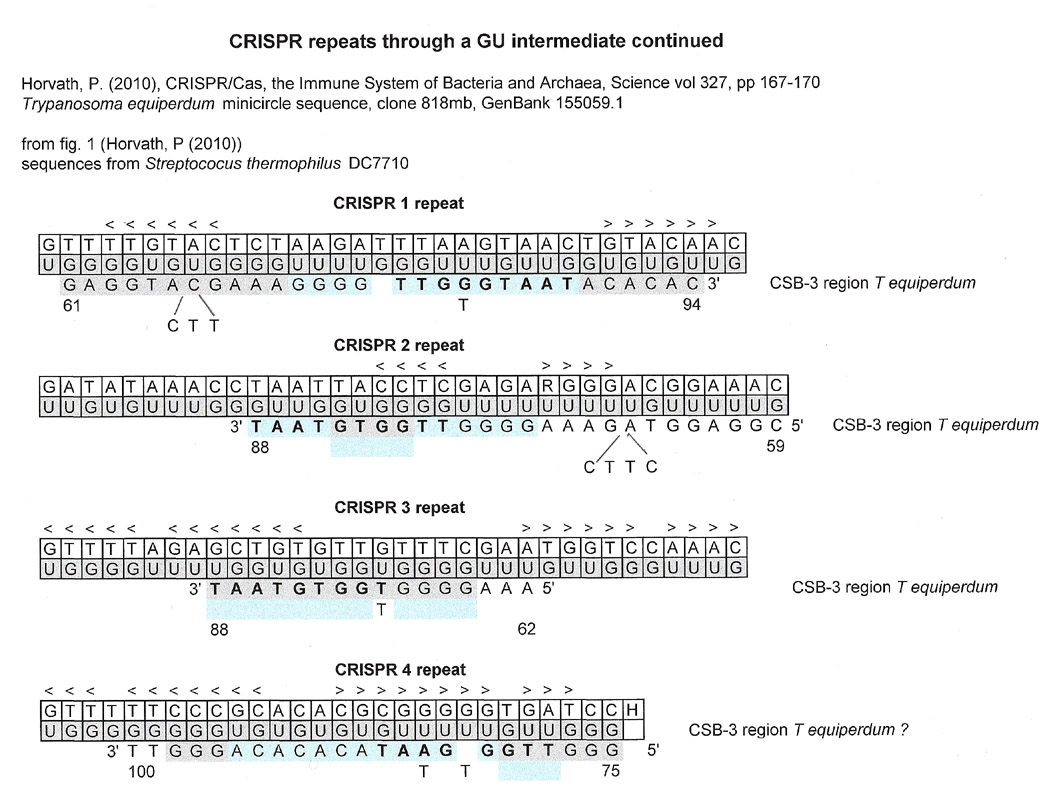
Below are CRISPR repeat sequences from two separate organisms, Syntrophus aciditrophicus and a strain of Streptococcus thermophilus. The repeats are all different, but all share a stem loop structure. The single-stranded loops do not coincide with the single stranded kinetoplastid gap sequence but nevertheless a linear match for the CSB-3 region can be found in all.
The fact that there is an alignment match does not mean any causal relationship between the CSB-3 region sequence and the CRISPR repeat.
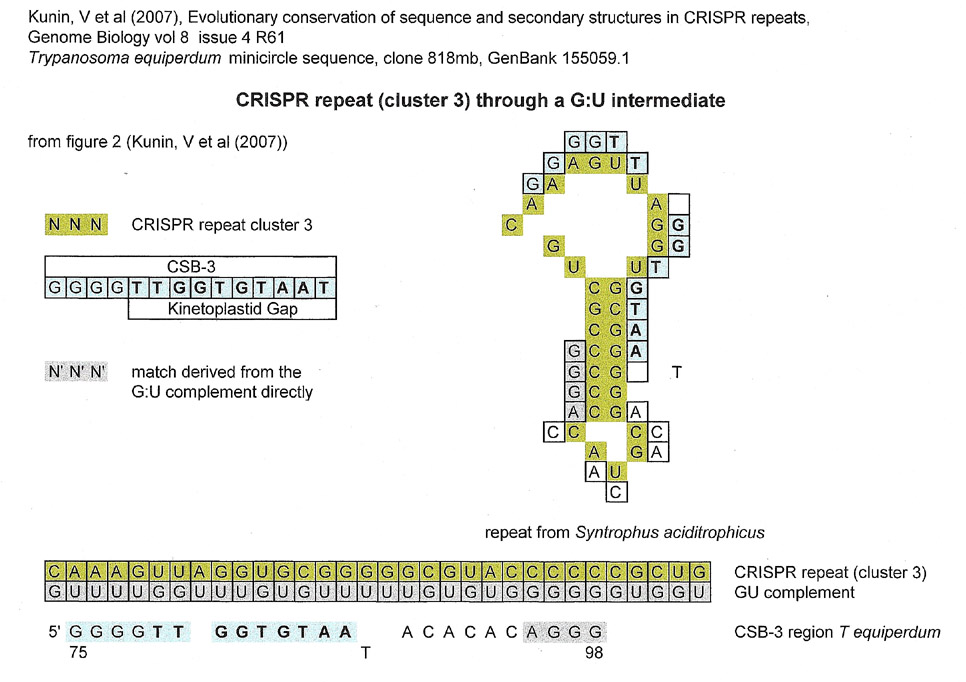
< < < < < < >>>>>>> - stem region sequence complements for the results below
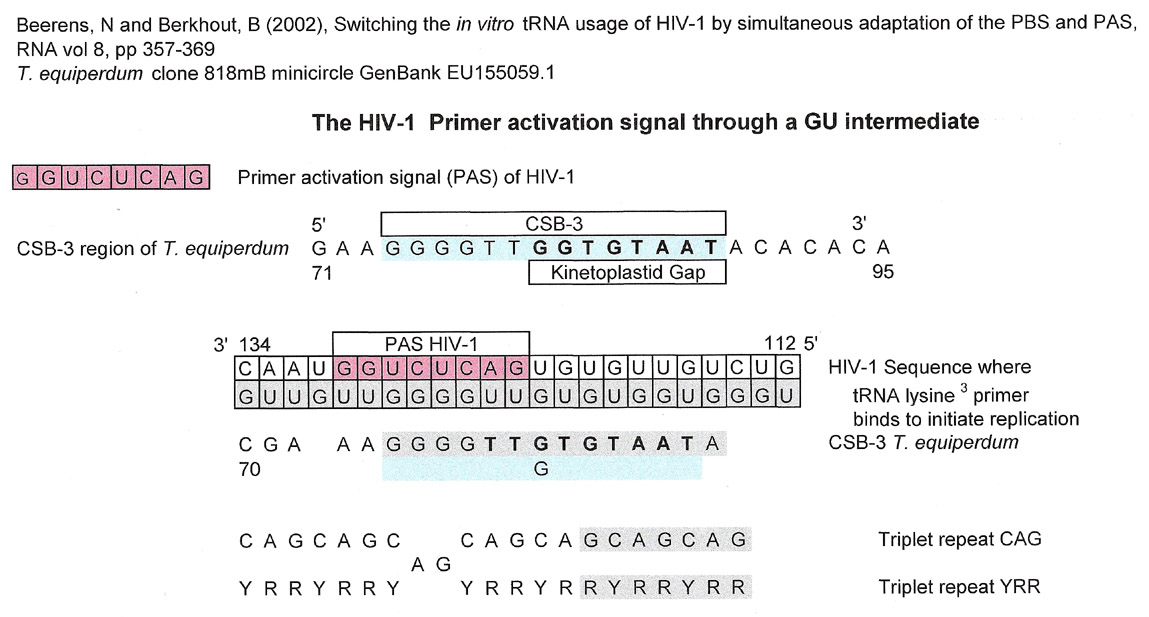
Beerens, N and Berkhout, B (2002) illustrate a series of tRNA primers (page 366) that bind at two locations to their respective retroviruses. The two sites are the Primer Activation Site (PAS) and the Primer Binding Site (PBS). The PAS region of HIV-1 where the anti-PAS sequence of tRNA lysine 3 binds is shown below. Although the match is not aligned with the kinetoplastid gap sequence and is at the end of CSB-3, the graphic was kept because of the match to the triplet repeat.
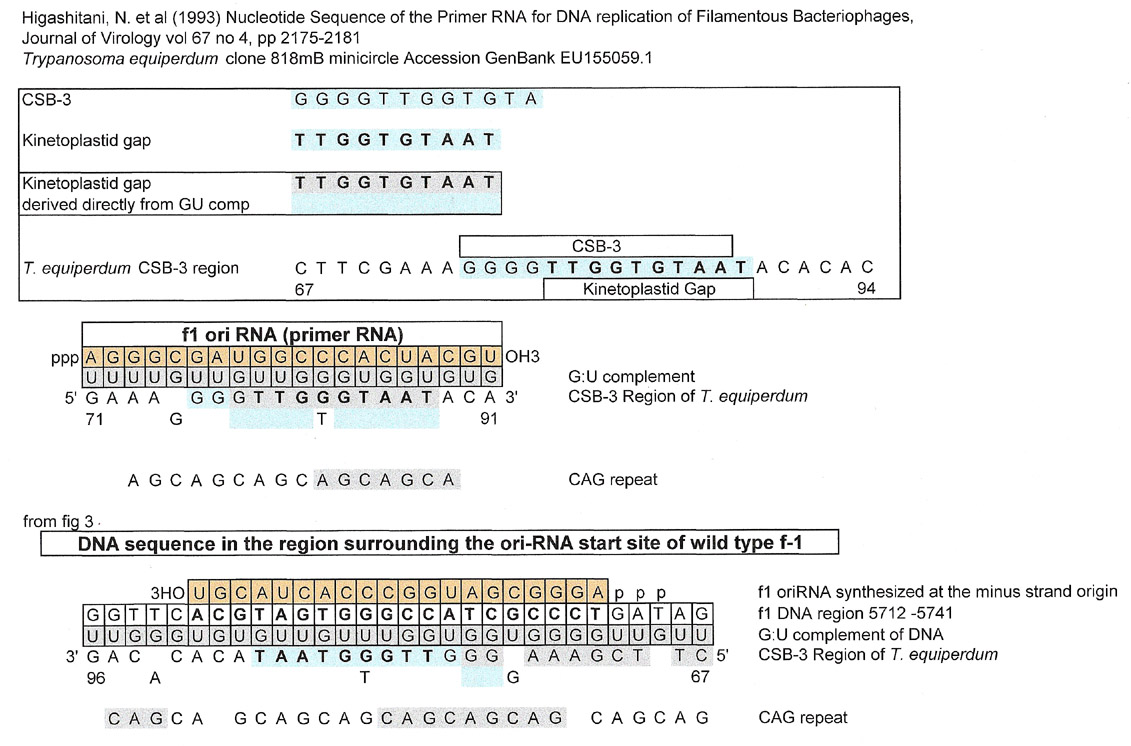
Primer RNA for DNA replication of a Filamentous Bacteriophage fi
The paper below is fairly complicated and describes the various mutations found in the TAU gene on chromosome 17 that lead to aberrant splicing producing an "incorrect" TAU protein that in turn becomes the filamentous inclusions found in FTDP-17. (Frontemporal dementia with parkinsonism chromosome 17 type).
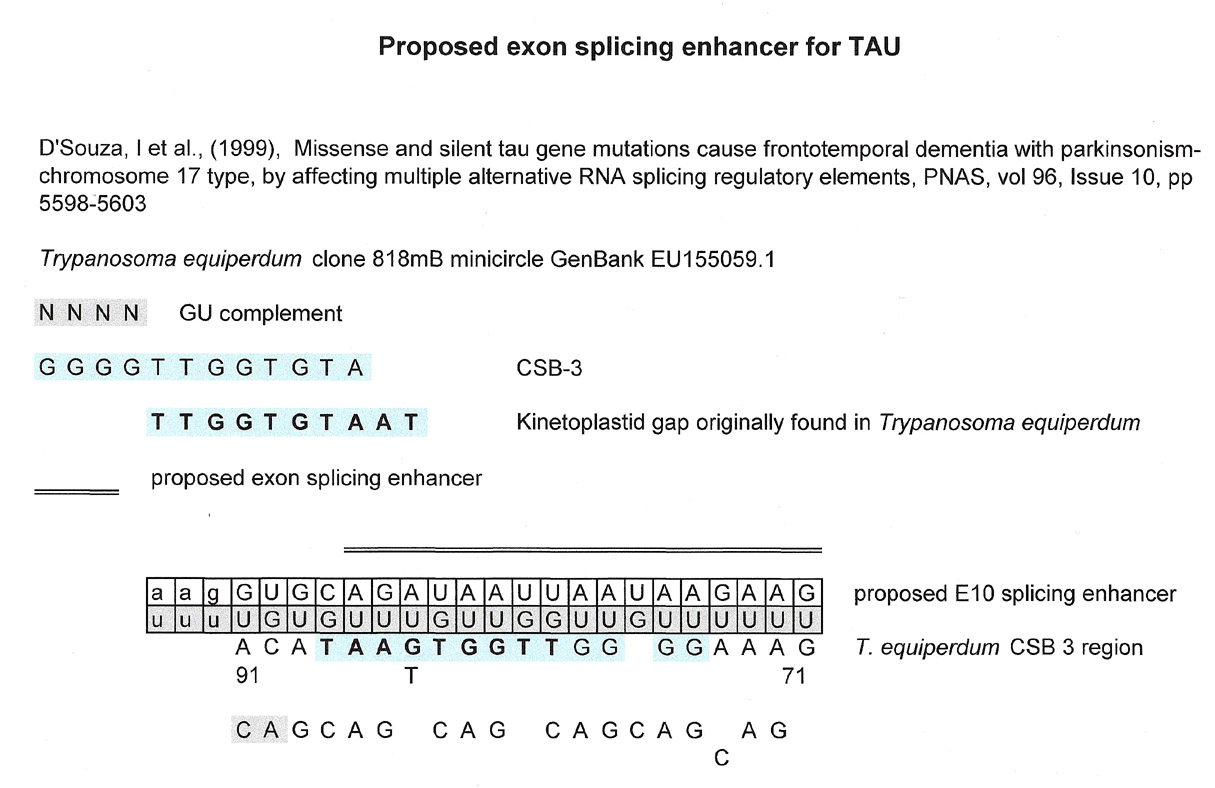
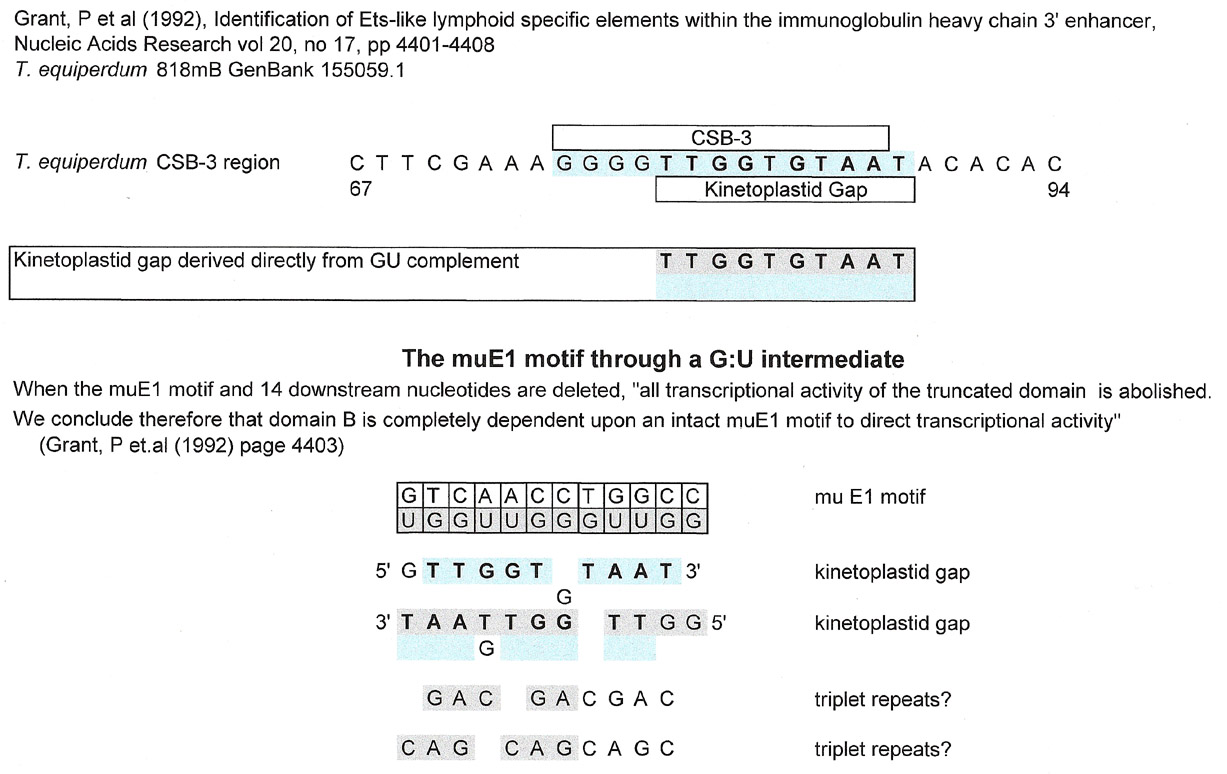
The graphic below is from a paper that studies, through sequence analysis, using a reporter system, the three functional domains of the 3' enhancer of the IgH. You are referred to the paper for further explanation.
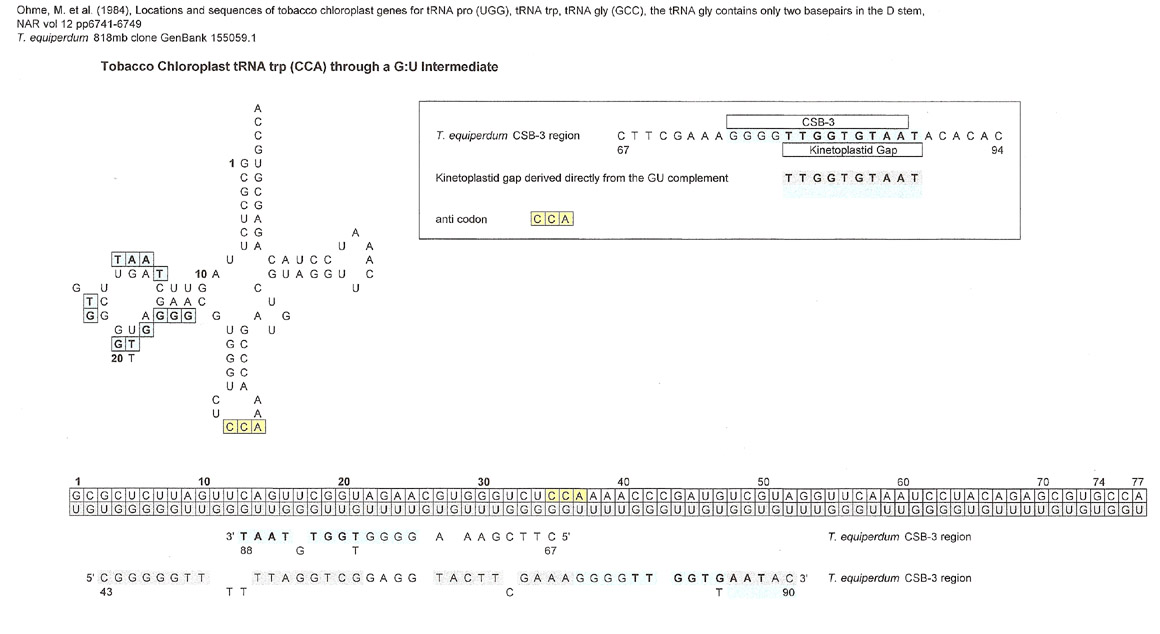
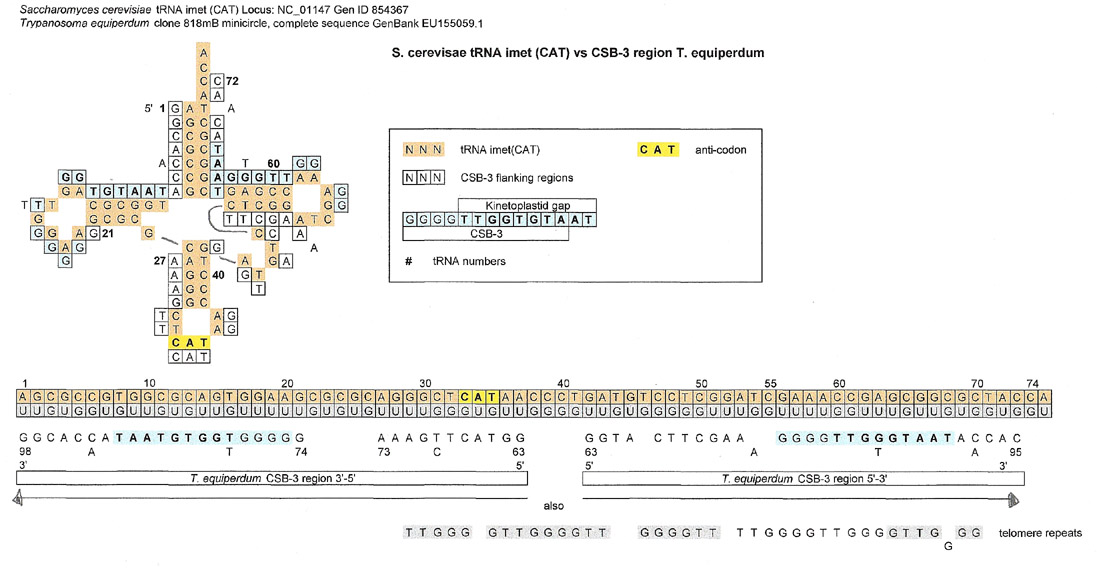
A few tRNAs were searched for matches, but only three results will be displayed.
The tRNA below, from S. cerevisae, is interesting because two symmetrical matches can be made for what might be a 5' side and a 3' side of the molecule. It is noteworthy that the symmetry is around the 37-38 region of the tRNA. This brought to mind the many pre-tRNAs in archaea that have introns spliced out at 37/38 and 38/39.(Marck, C. and Grosjean, H. (2003)). Unfortunately I was unable to find if the S. cerevisiae reported below originally contained an intron and was processed. The sequence was verified using http://trna.bioinf.uni-leipzig.de
Although no evidence of splicing out of an intron was found for the above tRNA, splicing and ligation was found in the processing of pre-tRNA leucine (UAA) of Cyanidioschyzon merolae. (Soma et al. (2007) See graphic below.) There is a match for CSB-3 at the 20/21 junction. The authors posit that the pre-tRNA sequence is transcribed with the 3' half of the molecule 5' to the 5' half, then the molecule is circularized and then processed so the finished tRNA has the 5' half (blue letters) in the usual place; 5' to the 3' half. (red letters). The 20/21 junction in the finished tRNA is not a join where an intron was excised out, but where the whole molecule was clipped out of linear RNA, The 20/21 join is in the D loop of the processed tRNA. The numbers are for the final processed tRNA.
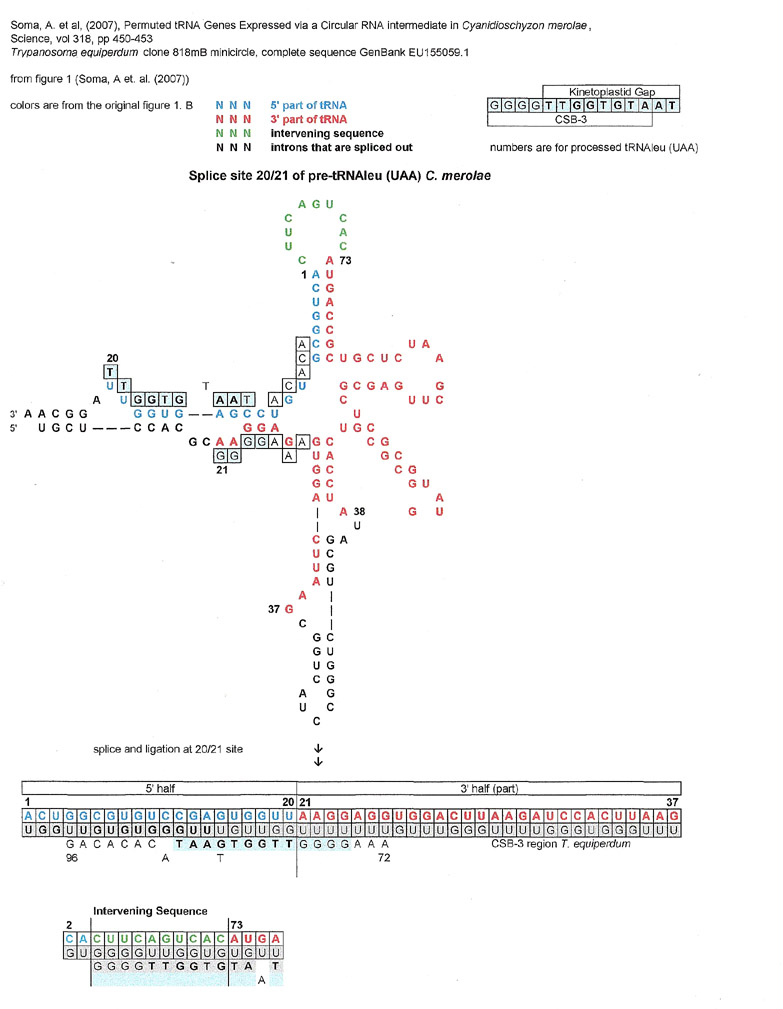
A quick note, through a GU intermediate, the intervening sequence loop is a very close complement match for the single stranded loop of the CRISPR repeat from Syntrophus aciditrophicus described above under CRISPRS
| C | A | C | U | U | C | A | G | U | C | A | C | A | U |
pre-tRNA leucine (UAA) C.merolae |
|||||
| 1 |
Intervening Sequence Loop
|
73 | |||||||||||||||||
| G | U | G | G | G | G | U | U | G | G | U | G | U | G |
GU Intermediate |
|||||
| C | A | A | A | G | U | U | G | G | U | G | C | G |
CRISPR Repeat Sequence |
||||||
| A | |||||||||||||||||||
|
CRISPR Repeat Single stranded loop
|
|||||||||||||||||||
Group II intron DNA insertion site
(endonuclease binding site)

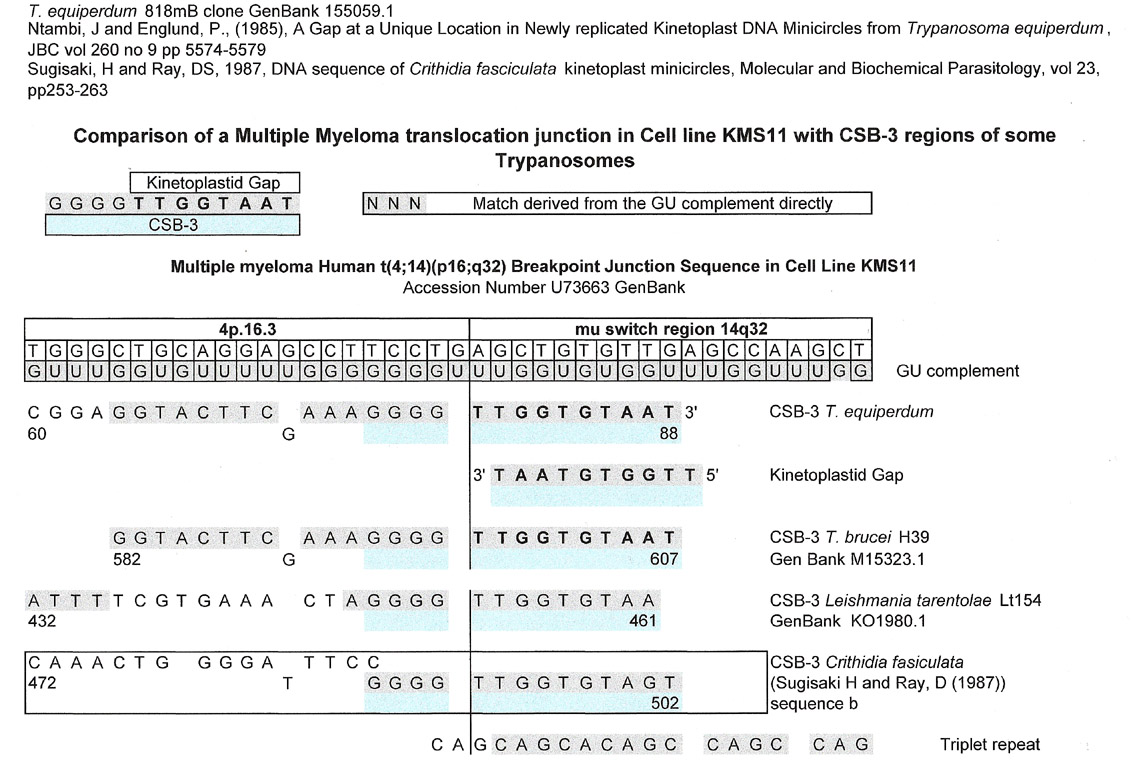
As can be seen below, there are a number of trypanosome CSB-3 matches for a multiple myeloma cell line (KMS 11) at its translocation junction. The kinetoplastid gap is a match for the IgH mu switch region 14q32. Notably, the junction sequence is also a match for the CSB-3 regions of strains of T. brucei, L. tarentolae, and C. fasciculata.
Using a G:U intermediate, one can see a definite relationship amongst the kinetoplast sequences that match the 4p.16.3 region that might not be seen if one tried to make homologies along the sequence of interest. The fact that a match can be made for all four kinetoplasts from both the sequence of interest (4p.16.3) and its G:U complement directly, shows that the kinetoplast sequences may have originated from the same "ancient" sequence....but one cannot prove this. These results also may imply that the actual original CSB-3 region may be the result of a splice and ligation as in the C. merolae tRNA shown above.
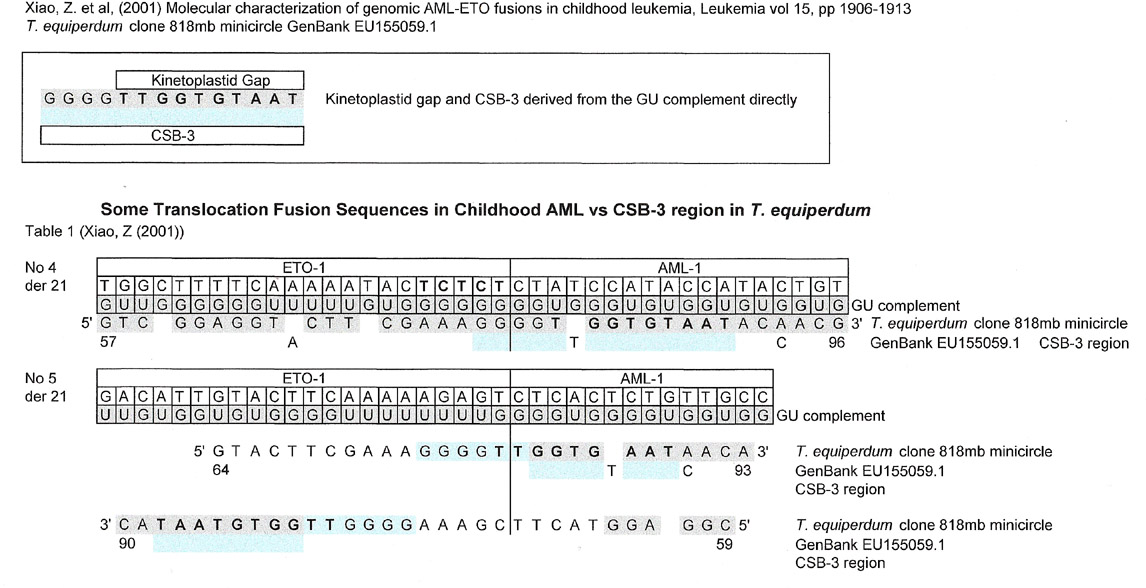
The match below for two ETO-1/AML fusions in childhood leukemia are good because the first (der21, #4) shows a match to the CSB-3 region of T. equiperdum derived solely from the G:U intermediate itself and actually spans the complete sequence reported in Xiao, Z. et al's paper. The second junction sequence (der21 #5) the matching 5' to 3' sequence is split, almost at the junction, between the sequence of interest ETO and the GU complement of AML-1.
Telomere Repeats, human antibody mu switch region

There are many matches for microRNAs but the examples below were chosen because they were derived from experiments. According to the authors, RRAS2 is strongly expressed in skin cancer and is a target of miR-23b. Of note in the match below is that the GU complement of the RRAS2 target sequence has many more complementary bases than the complement reported by the authors.

The miRNA precursor was included below simply to show that the match extended beyond the processed miRNA.
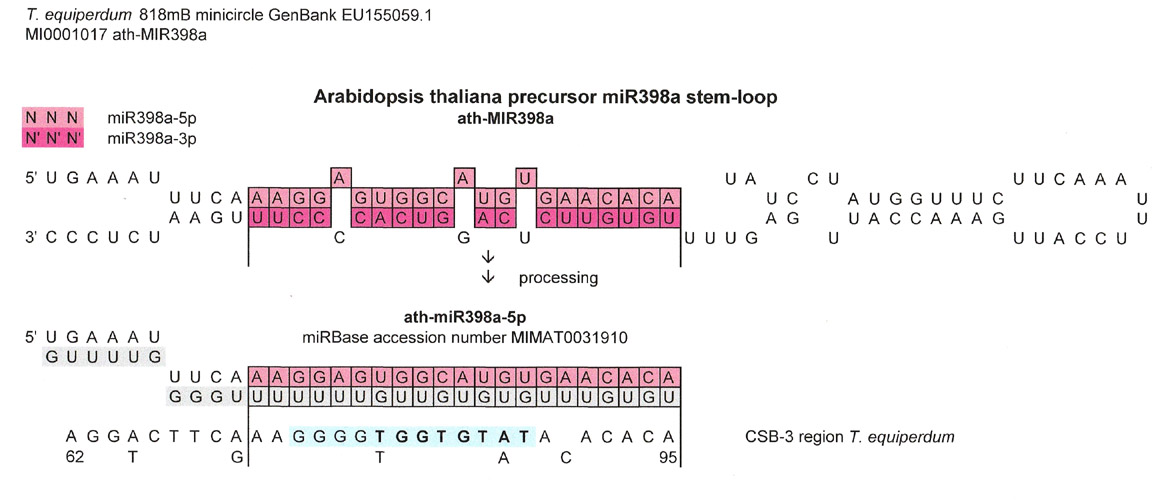
Even though this installment of Snippets is about the various matches to the universal kinetoplastid gap and CSB-3 region, I am including a finding of triplet repeats to make some quick points.
Like one of those dolls that has many layers, some sequences may have many matches, not only 5'-3' or 3'-5' or from both its plus strand and its GU complement directly but from sequences, for want of a better word, "underneath". Assuming that all these matches reflect something real and are not wishful "best-fit" thinking, it says something about how these sequences, billions of years old, might have been "used" or evolved for various biological mechanisms in various organisms or at various times.
It is unclear whether this implies some basal sequence, or in the case of triplet repeats, some reiterative mechanism.
Now for some geek fun. The triplet repeats found in the miRNAs reported below are significant because this author found two major groups. Those derived from the first group, a mix of YRR/RRY and a second group, YYYRRR/RRRYYY. The first group can also become RYR/YRY through a "frameshift" of one nucleotide. So, for instance (UGG)n can also be (ATG)n through a G:U complement.
| U | G | G | U | G | G | U | G | G | U | G | G |
| G | U | U | G | U | U | G | U | U | G | U | U |
| C | A | G | C | A | G | C | A | G | C | A | G |
| T | A | A | T | A | A | T | A | A | T | A | A |
| T | G | A | T | G | A | T | G | A | T | G | A |
| A | C | C | A | C | C | A | C | C | A | C | C |
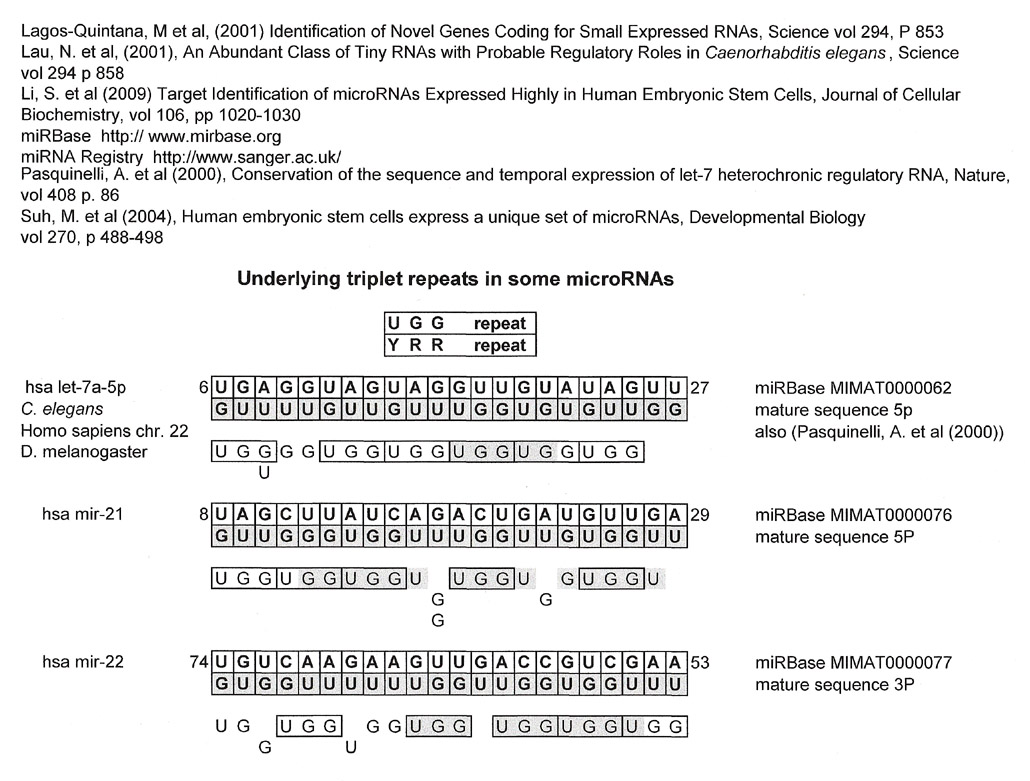

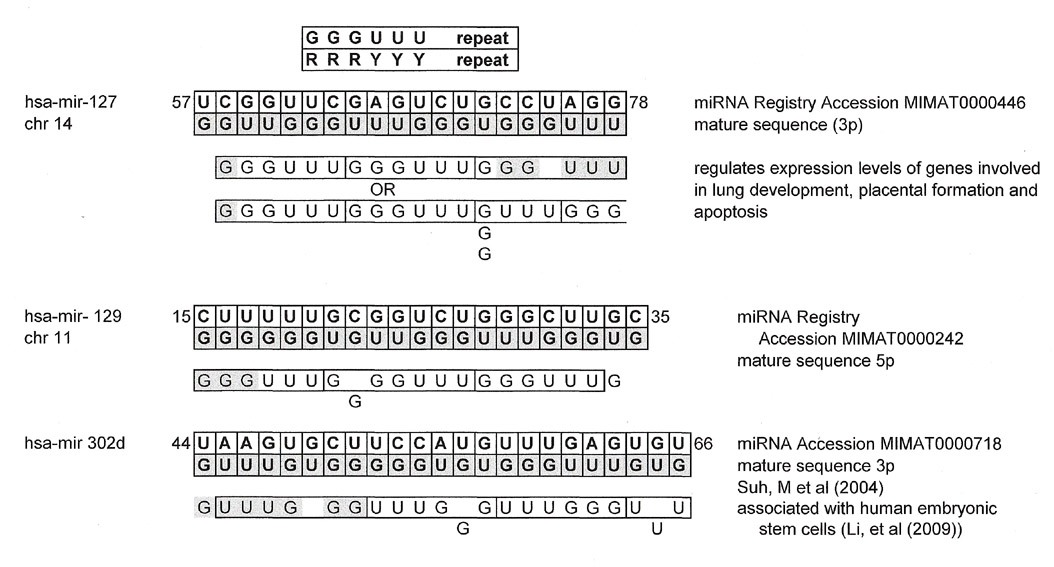
Comments on Results
One has to understand that you can find more than one match for any given sequence of interest. Most sequence analyses up until now were to find homologies and if found, there was only one homology per sequence (give or take a base or two). Using a G:U complement to make matches generates more than one match per sequence. One has to get used to generating results that are not very "neat".
What is described above in this installment of Snippets is for non-coding regions and not for open reading frames which makes the finding of underlying triplet repeats, especially two sets, as in the miRNAs, very intriguing.
One more point. CSB-3 is one of three regions in kinetoplastid minicircles that are conserved, although the others, CSB-2 and CSB-1, are less stringently conserved. CSB-1 and CSB-2 are separated by roughly 28 base pairs, and CSB-2 and CSB-3 are separated on average by about 47 pairs in a minicircle. (Ray, D (1989)). I have found other matches where CSB-1,2 and 3 have no intervening bases between themselves (for a further installment of Snippets).
I also found this lack of separation of conserved sequences in hammerhead ribozymes, as reported in Snippets 19. The usual hammerhead ribozyme has helices between its conserved parts, and in the case of some miRNAs, matches could be made between an miRNA and a hammerhead ribozyme that had no helices.
One cannot make any sweeping generalizations for if these many matches represent something real on some sort of evolutionary scale, then they generate more questions and an analysis of them has the same drawbacks that, say, a paleontological approach has. These sequences are billions of years old and while I have found things here or there (in this case matches) it is very difficult to draw conclusions about their evolution or correlation to known biological mechanisms without more analysis.
The author would like to thank the New York Public Library for accepting her as an "independent scholar" in its MaRLI program that allows her to have access to a consortium of libraries and their e-journal databases.
References
Abu-Einell, et. al (1999) Universal Minicircle Sequence Binding Protein, a Sequence Specific binding Protein that Recognizes the Two Replication Origins of the Kinetoplast DNA Minicircle, The Journal of Biological Chemistry, vol 274, pp 13419-13425.
Beerens, N and Berkhout, B (2002), Switching the in vitro tRNA usage of HIV-1 by simultaneous adaptation of the PBS and PAS, RNA, vol 8, pp357-369
Caoile, A and Stern, D (1997), A conserved core element is functionally important for maize mitochondrial promoter activity in vitro, Nucleic Acids Research, vol 25, pp 4055-4060.
D'Souza, I et. al., (1999), Missense and silent tau gene mutations cause frontotemporal dementia with parkinsonism-chromosome 17 type, by affecting multiple alternative RNA splicing regulatory elements, PNAS, vol 96, issue 10, pp 5598-5603.
Grant, P. et. al., (1992), Identification of Ets-like lymphoid specific elements within the immunoglobulin heavy chain 3' enhancer, Nucleic Acids Research, vol 20, no 17, pp 4401-4408
Guo, H. et. al., (1997), Group II intron endonucleases use both RNA and protein subunits for recognition of specific sequences in double-stranded DNA, The EMBO Journal, vol 16, no 22 pp 6835-6848.
Helmann, J, (1995) Compilation and analysis of Bacillus subtilis s A dependent promoter sequences: evidence for extended contact between RNA polymerase and upstream promoter DNA, Nucleic Acids Research, vol 23, pp2351-2360
Higashitani, N. et. al., (1993) Nucleotide Sequence of the Primer RNA for DNA Replication of Filamentous Bacteriophages, Journal of Virology, vol 67, no 4, pp 2175-2181.
Horvath, P., (2010), CRISPR/Cas, the Immune System of Bacteria and Archaea, Science vol 327, pp167-170
Inouye, S. et. al., (1986), Complete nucleotide sequences and genome organization of a Drosohpila transposable genetic element 297, European J Biochem, vol 154, no 2, pp 417-425.
Kraemer, A. et. al., (2013) UVA and UVB Irradiation Differentially regulate microRNA Expression in Human Primary Keratinocytes, PLoS one, vol 8, issue 12 e83392
Kunin, V. et. al., (2007), Evolutionary conservation of sequence and secondary structures in CRISPR repeats, Genome Biology, vol 8, issue 4 R61
Lagos-Quintana, M. et. al, (2001), Identification of Novel Genes Coding for Small Expressed RNAs, Science vol 294, p. 853
Lau, N. et. al., (2001), An Abundant Class of Tiny RNAs with Probable Regulatory Roles in Caenorhabditis elegans, Science vol 294, p 858
Li, S. et. al., (2009), Target Identification of microRNAs Expressed Highly in Human Embryonic Stem Cells, Journal of Cellular Biochemistry, vol 106, pp 1020-1030
Marck, C. and Grosjean, H (2003), Identification of BHB splicing motifs in intron-containing tRNAs from 18 archaea: evolutionary implications, RNA vol 9 pp 1516-1531.
Ntambi, JM and Englund, PT (1985) A Gap at a Unique Location in Newly Replicated Kinetoplast DNA Minicircles from Trypanosoma equiperdum, The Journal of Biological Chemistry, vol 260, pp 5574-5579
microRNA sequences from mirBase (http://www.mirbase.org) or miRNA registry (http://www.sanger.ac.uk) if not referenced to specific articles
Ntambi, JM et al (1986), Ribonucleotides Associated with a Gap in Newly Replicated Kinetoplast DNA Minicircles from Trypanosoma equiperdum., The Journal of Biological Chemistry, vol 261, pp 11890-11895
Ohme, M. et. al., (1984), Locations and sequences of tobacco chloroplast genes for tRNA pro (UGG), tRNA trp, tRNA gly (GCC), the tRNA gly contains two base pairs in the D stem, Nucleic Acids Research, vol 12, pp 6741-6749.
Pasquinelli, A. et. al., (2000), Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA, Nature, vol 408, p. 86
Ray, D., (1989), Conserved Sequence Blocks in Kinetoplast Minicircles from Diverse Species of Trypanosomes, Molecular and Cellular Biology, vol 9, no 3, pp 1365-1367
Saccharomyces cerevisiae tRNA imet (CAT) NC_00147.6 or NC_00137.3, or NC_001148.4 (there are four on different chromosomes
Sugisaki, H and Ray, DS, (1987). DNA sequence of Crithidia fasciculata kinetoplast minicircles, Molecular and Biochemical Parasitology, vol 23, pp 253-263
Suh, M. et. al. (2004), Human embryonic stem cells express a unique set of microRNAs, Developmental Biology, vol 270, pp 488-498
Trypanosoma equiperdum clone 818mB minicircle sequence GenBank EU155059.1
Vilkova, M. et. al., (2005), The Evolutionary origin of insect telomeric repeats (TTAGG)n, Chromosome Research, vol 13, pp 145-156.
Voskuil, MI and Chambliss, GH, (1998), The -16 region of Bacillus subtilis and other gram-positive bacterial promoters, Nucleic Acids Research, vol 26, no 15, pp3584-3590.
Xiao, Z. et. al., (2001), Molecular characterization of genomic AML-ETO fusions in childhood leukemia, Leukemia, vol 15, pp 1906-1913.